
In this example, we are interested in impementing a complex biological model inpired from the work of Bottino et al. This model simulates the sperm motility of the nematod Ascaris suum.
The Ascaris suum sperm cell crawls using a lamellipodial protusion, adhesion, retraction cycle. The chemical mechanisms of motility are located in the front of the cell called lamellipodium. In this model, the system corresponds to the lamellipodium membrane stuck to the matrix surrounding the cell. First, a fibrous polymerization occurs at the leading edge of the cell creating protusions. These protusions push the cell membrane forward. Then, some elastic energy is stored in the created fibrous gel. During the adhesion step, the protusions stick the matrix with a traction process that makes the cell body traveling. The fibrous gel undergoes a contraction. The final step occurs near the boundary between the lamellipodium and the rest of the cell where the depolymerization of the fibrous gel causes the deadhesion of the membrane. As a consequence, the stored energy is released to pull the cell body forward. This mechanics is moderated by a pH gradient.
Bottino describes this process within continuous differential equations. They correspond to the elastic and tensile stress in the membrane fixed to the extracellular matrix, and to pH distribution to deal with the pH dependence. In a second step, these equations are translated into a discrete form through a finite element model. The membrane is divided with a Delaunay graphe.
The translation of this discrete membrane using MGS topological collections and transformations is straightforward.
A node of the Delaunay graph is a record composed by 8 fields :
Fields px and py represent the node position in space, vx and vy its velocity, and H and pH the protons concentration and the pH. One defines one elements of type Node, some predicates to determine the exact type of the node.
The Delaunay graph is described by:
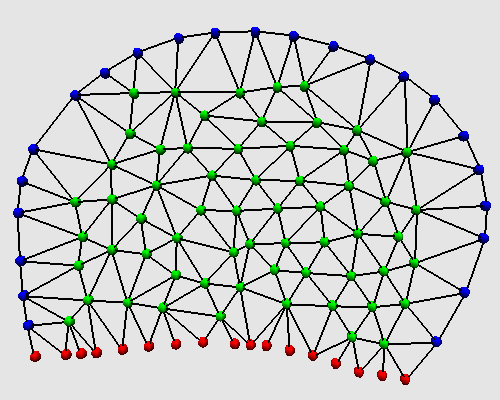
The polymerization and depolymerization processes are implemented by rewritting rules that
add or remove nodes:
where length returns the length of an edge between two nodes.
The differential equation of the pH are translated into a function that gives the new pH of a node depending
the pH of its neighbors:
The two first rules deal with de boundary conditions. The last one apply a diffusion equation between the internal nodes (Inode). We can note the using of the neighborfold primitive to compute the neighbors contribution.
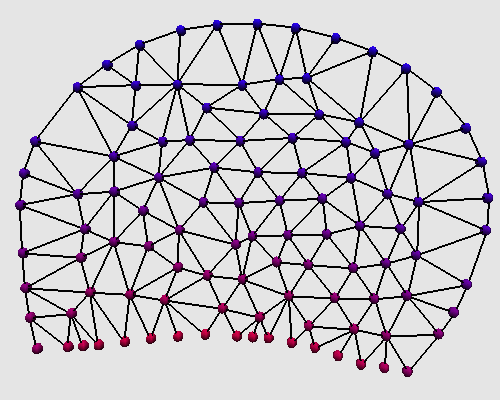
The transformation that computes mechanical forces applied on each nodes and that updates their postions,
is equivalent to
| Back
to top |
MGS examples index |
MGS home page |

Pictures, graphics and animations are licensed under a Creative Commons License.
|
|