This picture visualizes a Turing-like
reaction-diffusion process on a line of cells, but one of the two
chemicals is used as a trigger of the cell division. When its level is
above a given threshold, the cell divides into two cells and the concentration
in the two daughter cells is also divided. This process does not mimic an
identified natural phenomena but is used as an example of some kind of physico-chemical
process taking place in an expanding media.
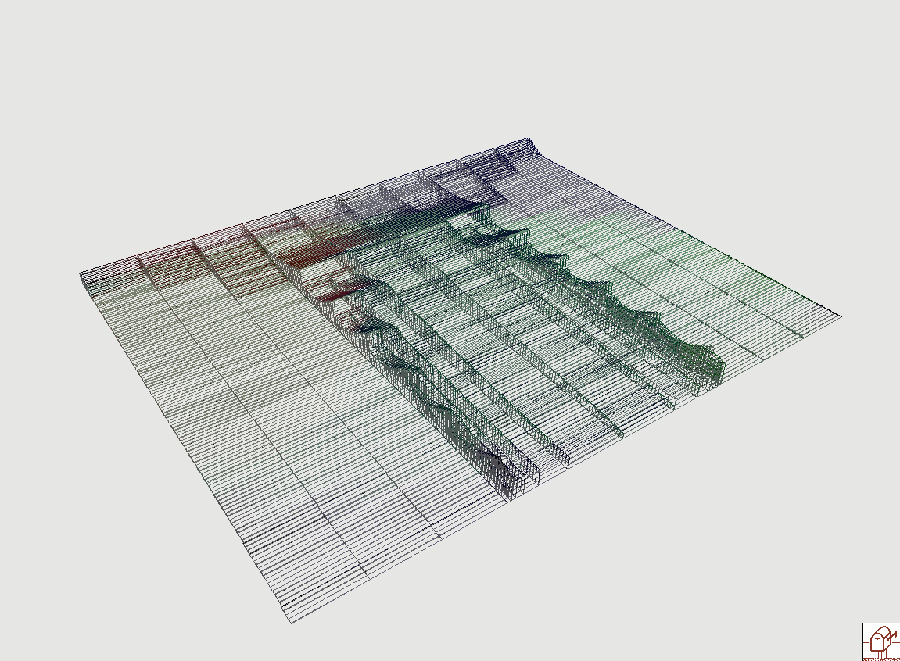
In the visualization below, the complete evolution of the filement is show. An elementary bloc corresponds to one cell, the line of cells is in the screen plane and the time is directed toward you. The height of a block is proportional to the quantity of the triggering chemicals present in the corresponding cell. The width of the daughter cells are divided by two, so a lineage corresponds to a parallel strip directed toward you. The data produced by the MGS simulation are written in a file using the TEOM format. This language is used to describe 3D scenes that are later rendered using the ImoView front-end (which is part of the MGS distribution).

|
specifying the Turing diffusion-reaction
and the cell division
|
|
| trans
init = { x => { a = 3.5 + random(1.0) -0.5, b = 4.0, beta = 12.0 + random(0.1) - 0.05, size = 16 }; };; NbCell := 20;; filement0 := init(iota(NbCell, ():seq));; |
The filemant of cell is represented
by a sequence of records; a record represents the state of one cell. Such a record has four fields: the fields a and b correspond to the concentration in morphogen a and b. The field beta is a constant used in the reaction-diffusion and that fluctuates a little randomly between the cells. The field size records the size of a cell. We start with 20 cells and the initial filament is made by applying the transformation init (which replaces each element x in a sequence by a record described as above) to a sequence made with the function iota. This function is reminiscent of the iota function in APL and builds a sequence of increasing integers. |
| rsp
:= 1.0/16.0;; diff1 := 0.25;; diff2 := 0.0625;; fun da(a, b, la, ra) = rsp * (16.0 - a*b) + diff1*(la + ra - 2.0*a);; fun db(a, b, beta, lb, rb) = rsp*(a*b - b - beta) + diff2*(lb + rb - 2.0*b);; |
Some constant parameters of
the reaction-diffusion process are defined. Function da and db computes the changes in morphogens a and b from the actual concentration of a cell and the concentrations of the left and right neighbors. |
| trans TuringSplit = { x / x.b > 8 => { a = x.a/2, b = x.b/2, beta = x.beta, size = x.size/2}, { a = x.a/2, b = x.b/2, beta = x.beta, size = x.size - x.size/2}; x / leftq(x) & rightq(x) => { a = x.a + da(x.a, x.b, left(x).a, right(x).a), b = max(0.0, x.b + db(x.a, x.b, x.beta, left(x).b, right(x).b)), beta = x.beta, size = x.size }; x / ~leftq(x) => { a = x.a + da(x.a, x.b, 0, right(x).a), b = max(0.0, x.b + db(x.a, x.b, x.beta, 0, right(x).b)), beta = x.beta, size = x.size }; x / ~rightq(x) => { a = x.a + da(x.a, x.b, left(x).a, 0), b = max(0.0, x.b + db(x.a, x.b, x.beta, left(x).b, 0)), beta = x.beta, size = x.size }; };; |
The transformation TuringSplit computes
the evolution of the filament. Each cell x is updated on the basis
of the following rules:
|
output of the TEOM description file |
|
| fun minimal(x)
= if (x <= 0) then 0.1 else x fi;; fun showX(x) = "Box { Size <1, " + minimal(x.b) + ", " + minimal(x.size) + "> Color <" + (0.99*(x.b/8)) + ", 0" + ", " + (0.99 - 0.99*(x.b/8)) + "> }"; |
A cell is represented as a
box. The function showX computes a string that defines this box
in the TEOM description language. TEOM is a language
used to defines 3D scenes that are then visualized by a viewer called ImoView.
The height, the width and the color of this box depends of the morphogens. |
| fun showBarre(barre, t,
tmax) = ( print_coll("tmp.turing.m", barre, showX, ("Grid1{ Axis <0,0,1> GridList [\n"), ",\n\t ", "] }"); if (t ~= tmax) then ("tmp.turing.m" << ",\n\n") else ("tmp.turing.m" << "\n\n") fi ); |
The function showBarre outpouts in the file
tmp.turing.m the description of the filament.
This description is build by the predefined function print_coll.
The argument of print_coll are the following:
print_coll(stdout, (\x.x), (1, 2, 3), "[", "#",
"]")
will print on the standard output:[1#2#3]
Here, the prefix used specify that the subsequent
box printed by showX must be aligned on a line.
|
| fun pre_show()
= ( "tmp.turing.m" << "Scaled{ Scale <0.1, 0.1, 0.1>\n" << " Geometry Grid1{ Axis<1,0,0> GridList[\n\n" );; |
At the begining of the TEOM
description file, we put some order to rescale the entire description. |
| fun post_show(n, c) = ( "tmp.turing.m" << "] }}\n"; close("tmp.turing.m"); system("imoview tmp.turing.m") );; |
At the end of the simulation, we close the TEOM
description file and we launch the ImoView visualization interface on this
file. |
| fun evol(barre,
t, tmax) = ( showBarre(barre, t, tmax); if (t < tmax) then evol(TuringSplit(barre), t+1, tmax) else barre fi ); |
This function iterates the
evolution of the filament (by applying the core tansformation TuringSplit)
and the output of each filament's state. |
| fun evolve(n) = ( pre_show(); evol(filament0, 0, n); post_show(n, NbCell) );; evolve(380);; |
|
| Back
to top |
MGS examples index |
MGS home page |

Pictures, graphics and animations are licensed under a Creative Commons License.
|
|