| Figure 1: description (taken from [Mes05]) of the regulation network of the CI gene of the lambda phage. The RNA-polymerase is in red, a DNA strand appears in yellow and the repressor monomers that combines into dimers are in blue. |
Stochastic simulation is of interest when the number of molecules and/or the time-step between each event of the modelized system is small. The lambda phage is a virus that infects the cells of the Escherichia coli bacteria. Once having infected the bacteria, the virus has two possible developmental pathways. It can either (1) replicate and lyse (dissolve) the host cell and release about 100 progeny or (2) integrate its DNA into the bacterial DNA and form a lysogen. We describe the modelization and simulation in MGS of this phenomenon using a stochastic rule application strategy following Gillespie's Stochastic Simulation Algorithm. A complete biological description of the problem can be found here for example.
In short, the virus that infects the bacteria has two possible outcomes: lyse or lysogeny. According to the local environment, one of the two outcomes is chosen, the decision being under control of a small region of the genome, two genes (CI and CRO) and two promotors. This regulation region is called the genetic switch. Each of thow two proteins CI and CRO will repress the synthesis of the other one and increase its own production. Following local conditions, one of the two protein will take advantage over the other and the system will enter in either the lyse or lysogeny phase.
|
Figure 1 represents a fragment (limited to the only description of CI) of the regulation network of the transcription of the lambda phage. The protein (stated in blue on the figure) is a repressor and takes place in the transcription process of CI, by activating or inhibiting the proces. This molecule (equation (1)) is able to dimerize and monomerize. Equation (8) states the degradation of the repressor. Equations (2)-(5) describe the bond of the repressor molecules on the DNA strand. When dimers are missing (or when only one or two molecules are present) on the DNA strand, the transcription of a molecule of CI is possible (equations (6) and (7)) while it becomes impossible when a third ones appears (equation (4)).
The translation of the network in chemical equations (the Ki correspond to kinetic constants) is:
![\begin{equationarray}[c]{lcl}](img110.png)
Below is given the result of seven simulations, after 2000
simulations steps, of the model (click
on each picture to enlarge the view), with an initial
state made of (#3
`CI, `D0, #10 `P):
The translation into an MGS program is straightforward. Each equation is translated into a rewrite rule and the value of the constant C is determined by biological experiments(see [ KN05]). We give here the MGS program that describe the laws for the behaviour of Cro and CI :
| trans
A = { // // Description of Cro // `Cro ={ C = 0.005 }=> <undef>; #2 `Cro ={ C = 0.01 }=> `Cro2; `Cro2 ={ C = 0.25 }=> #2 `Cro; `D0, `P ={ C = 0.05 }=> `D0, `P, `Cro; `D0, `Cro2 ={ C = 9.768 }=> `D'3; `D0, `Cro2 ={ C = 9.768 }=> `D'2; `D'3 ={ C = 29.77 }=> `D0, `Cro2; `D'3, `P ={ C = 0.05 }=> `D'3, `P, `Cro; `D'3, `Cro2 ={ C = 9.768 }=> `D'3D'2; `D'2 ={ C = 245.371 }=> `D'3, `Cro2; `D'2, `P ={ C = 0.05 }=> `D'2, `P, `Cro; `D'3D'2, `P ={ C = 0.05 }=> `D'3D'2, `P, `Cro; `D'2, `Cro2 ={ C = 9.768 }=> `D'1; `D'1 ={ C = 245.371 }=> `D'2, `Cro2; // // Description of CI // `CI ={ C = 0.0025 }=> <undef>; #2 `CI ={ C = 0.01 }=> `CI2; `CI2 ={ C = 0.25 }=> #2 `CI; `D0, `P ={ C = 0.005 }=> `D0, `P, `CI; `D0, `CI2 ={ C = 9.768 }=> `D1; `D0, `CI2 ={ C = 9.768 }=> `D2; `D1 ={ C = 15.557 }=> `D0, `CI2; `D1, `P ={ C = 0.005 }=> `D1, `P, `CI; `D1, `CI2 ={ C = 9.768 }=> `D1D2; `D1D2 ={ C = 15.557 }=> `D1, `CI2; `D1D2, `P ={ C = 0.086 }=> `D2, `P, `CI; `D2 ={ C = 399.225 }=> `D1, `CI2; `D2, `P ={ C = 0.086 }=> `D2, `P, `CI; `D2, `CI2 ={ C = 9.768 }=> `D3; `D3 ={ C = 2022.38 }=> `D2, `CI2 };; |
The
quations stated
above are directly translated into MGS rewrite rules. The
reaction constants are given as arguments to each rule specification in
the form of a C =
statement. |
| fun
inter(fic, coll) = ( let somme = count(`D0, coll) + count(`D1, coll) + count(`D2, coll) + count(`D3, coll) in (fic << 'iteration + "\t" + count(`Cro, coll) + "\t" + count(`Cro2, coll) + "\t" + count(`CI, coll) + "\t" + count(`CI2, coll) + "\t" + count(`D0, coll) + "\t" + count(`D1, coll) + "\t" + count(`D2, coll) + "\t" + count(`D3, coll) + "\t" + count(`D4, coll) + "\t" + count(`D'1, coll) + "\t" + count(`D'2, coll) + "\t" + somme + "\t" + count(`D'3, coll) + "\n"); coll);; |
Each value
that wants
to be plotted later has to be sent to a file (first argument of the
function). The current iteration, together with all the variables of
the simulation are written to the file. This function will be called after each transformation application onto the collection. |
| cpt := 0;; fun once() = A['fixpoint=(\x1,x2.('iteration >= 2000.0)), 'strategy = `gillespie, 'interlude = inter("/tmp/sortie.dat")](bagify(((#3 `CI), `D0, #10 `P)));; |
cpt
is a counter to memorize the current simulation count. After each
simulation, the current counter is printed and the result of the
simulation is plotted using gnuplot. The once function excutes a simulation (that stops after 2000 reactions, that is the argument of the `fixpoint option) and calls afterwards the inter function defined above. The strategy is specified using the `gillespie keyword of the 'strategy option. The initial state is a multisets made of 3 occurences of `CI, one `D0 and 10 occurences of `P. |
| fun
un_tour(x, y) = (system("rm -f /tmp/sortie.dat"); once(); close("/tmp/sortie.dat"); "/usr/bin/gnuplot" << "set output\n"; "/usr/bin/gnuplot" << "set term x11\n"; "/usr/bin/gnuplot" << "set xlabel \"Time\"\n"; "/usr/bin/gnuplot" << "set ylabel \"Population size\"\n"; "/usr/bin/gnuplot" << "set logsc y\n"; "/usr/bin/gnuplot" << "plot \"/tmp/sortie.dat\" using 1:3 title 'Cro2'with lines 2," + "\"/tmp/sortie.dat\" using 1:2 title 'Cro'with lines 3," + "\"/tmp/sortie.dat\" using 1:4 title 'CI'with lines 4," + "\"/tmp/sortie.dat\" using 1:4 title 'DNA'with lines 4," + "\"/tmp/sortie.dat\" using 1:5 title 'CI2'with lines 6\n"; "/usr/bin/gnuplot" << "replot\n"; sleep(1); cpt := cpt + 1; stdout << "Graph number " << cpt << "\n"; "/usr/bin/gnuplot" << "set output \"phage-" << cpt << ".eps\"\n"; "/usr/bin/gnuplot" << "set term postscript color solid\n"; "/usr/bin/gnuplot" << "replot\n"; sleep(1));; |
Function
un_tour
sends to gnuplot some instructions to plot the result of the current
simulation and prints out the number of the plot. The counter is then
incremented and a Postscript file is produced. |
| fold(un_tour,
0, 100);; |
Then the run of 100 experiments is launched. |
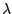 du
bactériophage
du
bactériophage  .
.  phage, Montpellier, april 2005.
phage, Montpellier, april 2005.| Back
to top |
MGS examples index |
MGS home page |

Pictures, graphics and animations are licensed under a Creative
Commons License.
|
|