the gif animation files are heavy: be patient while loading
The BLOB Computing project,
developped by F. Gruau at LRI,
represents an alternative to the current Von Neumann model, following
a biocomputation dogma.
The whole project idea is to try to capture the principles of
bio-computing system allowing massive parallelism. The model of
computation is based on the concept of self developing network of
compute nodes, the machine is a 2-D Cellular automaton grid whose
evolution rule is fixed and implemented by simplified physical laws. A
machine configuration represents idealized physical objects such as
membrane or particle gas. A central object called blob is the hardware
image of a compute node.
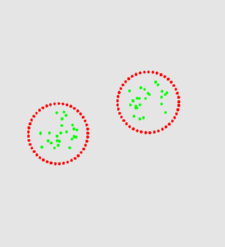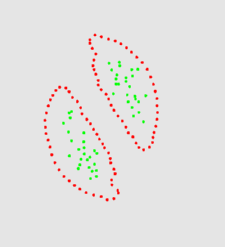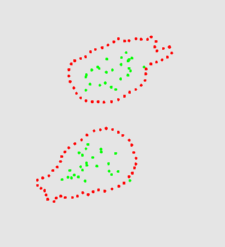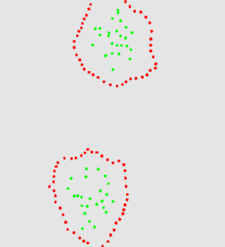
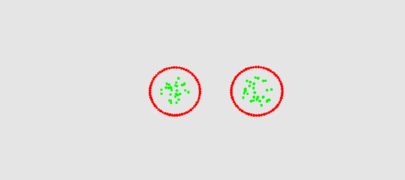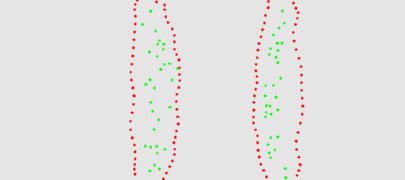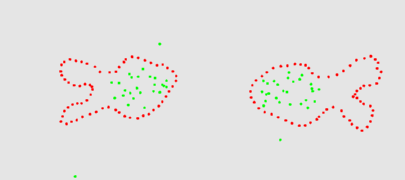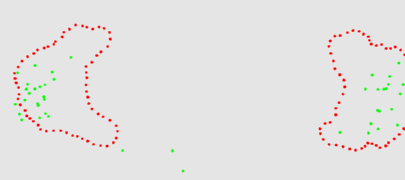
The two animation below shows the simulation of the collision of two blobs. Blobs are simulated in MGS using proxi. Proxy are a kind of topological collection where the neighborhood relationship is defined by a user predicate. Elements of the proxi used here are of two kinds: membrane nodes and molecule nodes. Membrane nodes are linked by a spring like force. All node exert a repulsive force. The loss of molecules in the movie is due to a too low attractive force between membrane nodes. Another version is based on the use of Delaunays. The two versions improve over an initial simulation coded as a cellular automaton. The simulation has been developped in MGS by Julien Cohen using a previous program developped by Antoine Spicher.


| Back
to top |
MGS examples index |
MGS home page |

Pictures, graphics and animations are licensed under a Creative Commons License.
|
|